Joseph Shiloach, Ph.D.

Professional Experience
- Director, Biotechnology Core Laboratory, NIDDK, 1979-2022
- Adjunct Professor, Johns Hopkins University, Department of Chemical and Biochemical Engineering
- Associate Professor, Georgetown University - Biochemistry and Molecular & Cellular Biology
- Research Associate, Tufts University, New England Enzyme Center, 1975-1978
- Ph.D., Hebrew University, Israel, 1975
Research Goal
The research purpose is to develop efficient methodologies for production of various biological materials, such as proteins and viruses, from different biological sources, especially microorganisms and mammalian cells. These biological products are being used for basic research, structural studies and clinical applications directed toward the development of effective new drugs and vaccines.
Current Research
The laboratory’s main interest is production, recovery and purification of biologicals from both prokaryotes and eukaryotes of native and recombinant sources. The work involves all aspects of this process including research, development, and production. In addition to research labs, the laboratory includes pilot production facility, well equipped with bioreactors of different sizes and variety of recovery and purification equipment. The bioreactors are used for propagation of biological systems such as mammalian cells, insect cells, bacteria, yeast, and fungi. The biological products made support clinical and structural studies.
We conduct research and process development on growth optimization, production, scale-up, and product-recovery processes. We concentrate on production and purification of proteins with emphasis on scaling up. The aim of our research studies is to achieve better understanding of the growth behavior and metabolism of both E. coli and several mammalian cells lines. The objective is to overcome specific difficulties in the growth and production process by modifying the properties of the producers. This work involves gene transcription and expression analysis as well as cell transfection and mutation. Our recent achievements are summarized below:
- We improved recombinant protein expression from HEK293 cells by knocking out the Caspase 8-associated protein 2 gene, which was originally identified by siRNA screening. This was following up by carful bioinformatic analysis that allow us to show that the mechanism involved the upregulation of the Cyclin dependent kinas Inhibitor 2A gene.
- We developed a novel knockout strategy to enhance recombinant protein expression in E. coli based on the phenomenon that recombinant protein expression triggers stress response associated with downregulation of key metabolic pathways. What we did is the following: instead of individually upregulating these downregulated genes or improving transcription rates by better vector design, we implemented a strategy that would block this stress response thereby ensuring a sustained level of protein expression.
- An improved expression of the repeating unit of the C difficile toxin A rARU (a potential vaccine against C. difficile) in E. coli was possible by restricting the oxygen supply (DO) to the producing culture. At restricted DO, rARU mRNA was 5-fold higher and the protein expression 27-fold higher compared with unrestricted DO. This strategy could improve the expression of recombinant proteins from anaerobic origin or those with cell wall binding profiles
- We identified genes involved in mammalian cell adhesion, which allows us to modify the way the cells grow. The gene Siat7e was overexpressed in the adherent MDCK cells and, as a result, the cells were able to grow in suspension. These cells were then utilized for the preparation of influenza vaccine replacing the current influenza vaccine production, which is based on using fertilized eggs.
- We identified microRNAs that are involved in controlling apoptosis onset in mammalian cells, allowing us to extend the growth phase of the cells and potentially enhance the production of biological compounds. Inhibition of the microRNA 466h increased the expression levels of anti-apoptotic genes and increased the cell viability.
- We improved glucose utilization by E. coli through the over-expression of small RNA. This was responsible for glucose transport, allowing the bacteria to grow to high density. Over expression of the small RNA sgrS in E. coliK strain reduced the level of the glucose transporter PTSG and allowed the bacteria to increase the glucose utilization yield, reduce acetate production, and to increase the bacteria concentration and recombinant protein production.
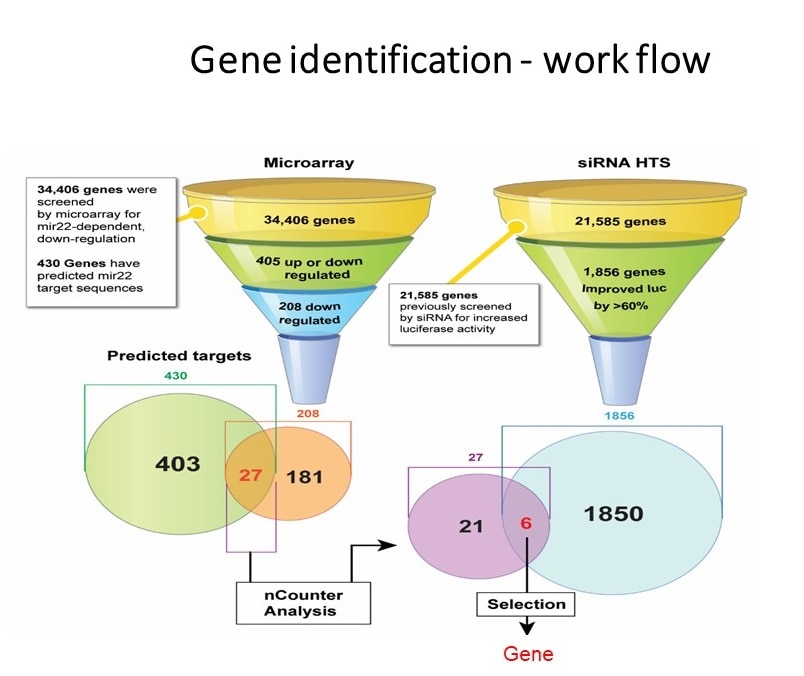
- By expanding our work related to noncoding RNAs we conducted High-Throughput screening of approximately 800 microRNA molecules. By careful analysis we identified 2 microRNAs that improved recombinant protein expression form HEK293 cell. This approach was initiated when large quantities of neurotensin receptor, hard to express proteins, were needed for structural studies.
- The high throughput screening approach of microRNA was expanded to screen more than 23000 genes by utilization human siRNAs library. After extensive analysis 10 genes whose inactivation was associated with improved recombinant protein expression were selected for further evaluation. The top candidate “Ornithine decarboxylase antizyme” (OAZ1) was knocked out using crisper technology to create an improved producing cell line.
- We developed a continuous production process of retroviral vector needed for adoptive T-cell Therapy. The process is based on growing the producing cells (PG13) on microcarriers in stirred tank bioreactor equipped with alternative tangential flow perfusion system. No detrimental effects on the specific viral vector titer or on the efficacy of the vector transducing the T- cells of several patients. Viral vector titer increased throughout the 11 days of the perfusion period. An indication that this method can be an efficient way to produce large quantities of active vector suitable for clinical use.
- Improved expression of protective antigen (PA) from Bacillus anthracis ames strain BH500. We developed newer approach to conduct extensive RNA seq analysis of samples from expressing and non-expressing Bacillus strain. As a result, we were able to identify key genes (not identified before) whose modified expression may improve the PA production.
Applying our Research
Improved methodologies for production and purification of biological compounds intended for clinical research will provide researchers with the materials needed for the evaluation and testing of potential new drugs and vaccines against various diseases.
Select Publications
- rAAV Production and Titration at the Microscale for High-Throughput Screening.
- Quan DN, Shiloach J.
- Hum Gene Ther (2022 Jan) 33:94-102. Abstract/Full Text
- Continuous production process of retroviral vector for adoptive T- cell therapy.
- Inwood S, Xu H, Black MA, Betenbaugh MJ, Feldman S, Shiloach J.
- Biochem Eng J (2018 Apr 15) 132:145-151. Abstract/Full Text
Research in Plain Language
My laboratory’s main interest is in developing ways to produce and isolate biological products from different biological sources such as mammalian cells, insect cells, bacteria, yeast, and fungi. Our work involves all aspects of this process, and in addition to a research lab the laboratory includes a pilot production facility to handle production and recovery from the various sources. The biological products we make are used by scientists involved with other research activities.
Research Images