About Our Research
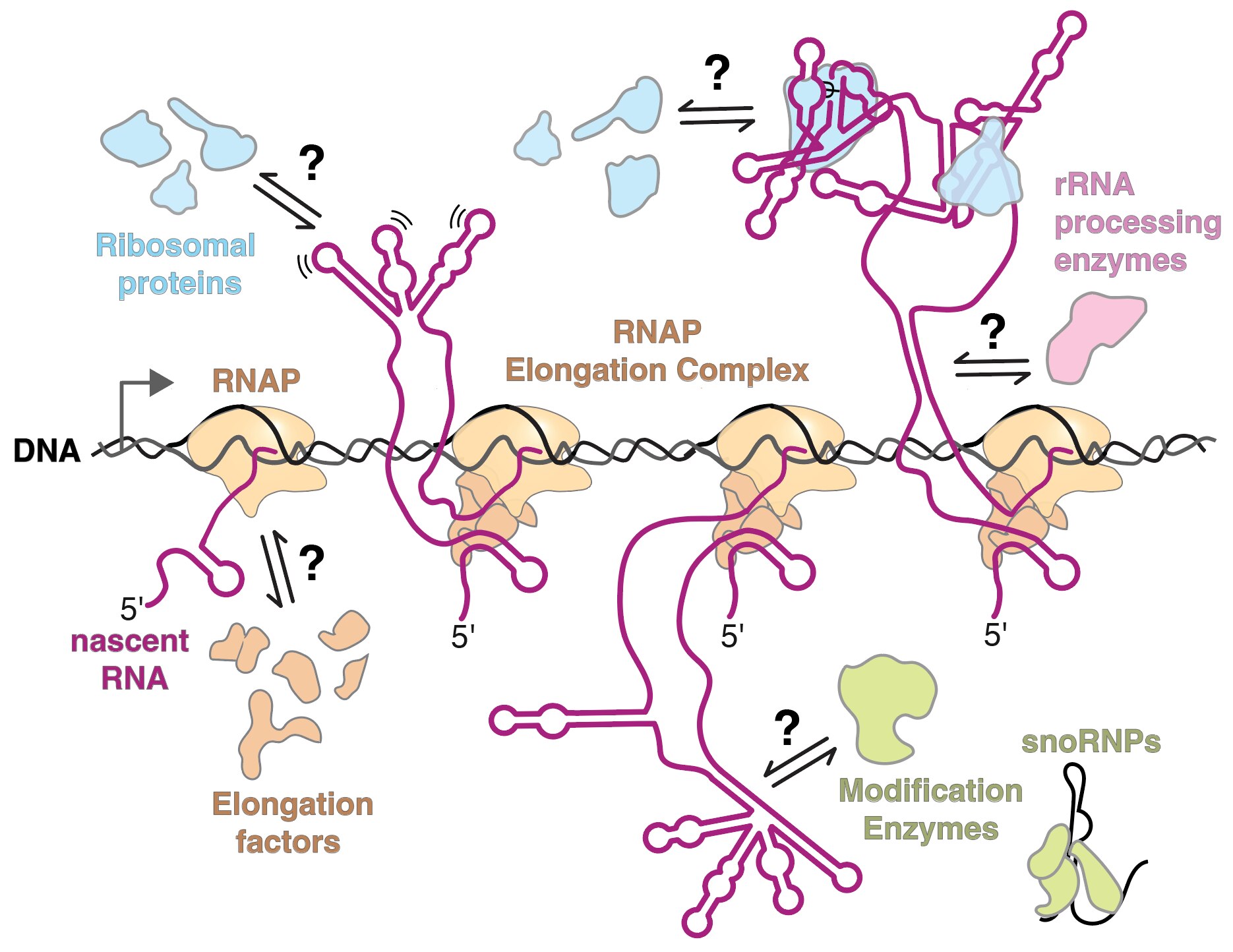
Ribonucleoproteins (RNP) are ubiquitous cellular workhorses involved in every step of gene expression, yet the underlying molecular mechanisms that cells use to assemble RNPs are poorly understood. For proper RNP assembly, an RNA must be accurately synthesized, modified, processed, folded, and bound by RNA-binding proteins. However, one challenge in RNP assembly is that it often occurs simultaneously with transcription of an RNA. Productive steps in RNP synthesis must compete with non-productive events that can occur during transcription, such as the formation of aberrant RNA structures as the RNA emerges from RNA polymerase. If the RNA forms an incorrect structure, then protein binding, processing, and modification may be delayed or inhibited altogether.
In the cell, RNPs are made exceptionally fast and with high fidelity in order to carry out essential steps in gene expression. For example, thousands of ribosomes – the RNP responsible for protein synthesis – are made every minute in a growing human cell in order to meet the demands for protein synthesis. Problems in ribosome assembly are extremely detrimental to the cell, with genetic defects in ribosomal components leading to diseases called ribosomopathies and dysregulation of ribosome assembly being associated with many cancers.
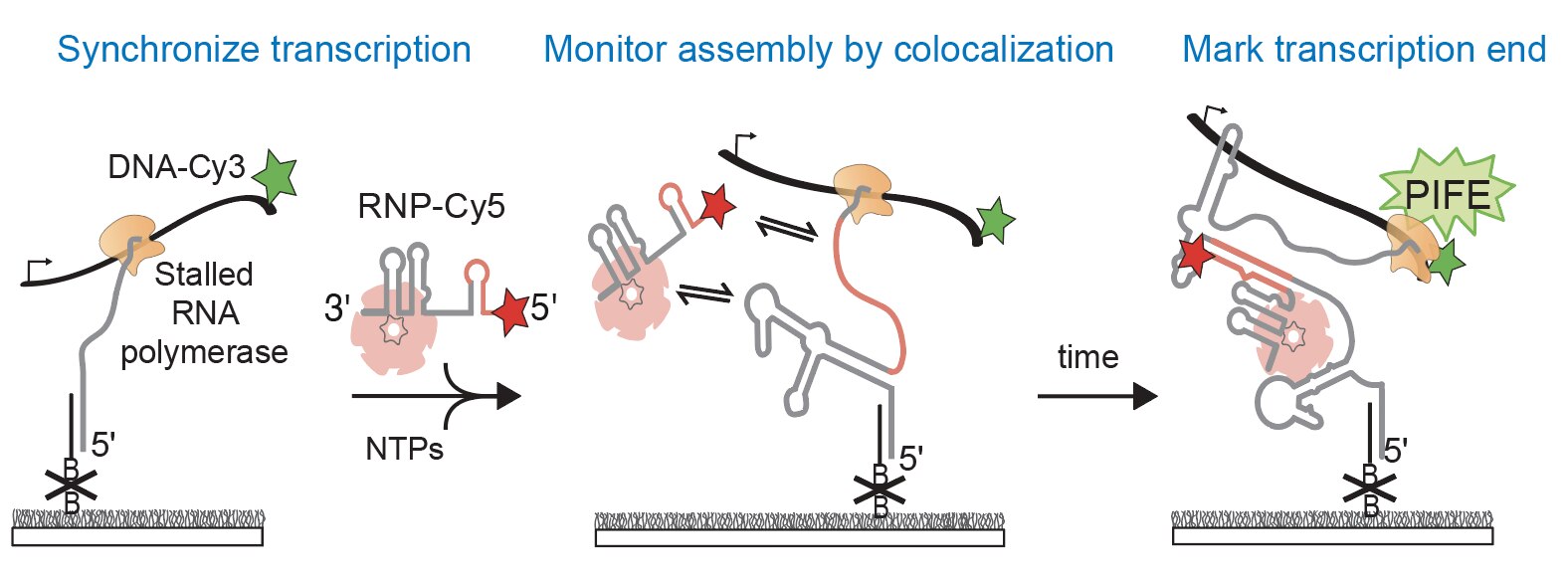
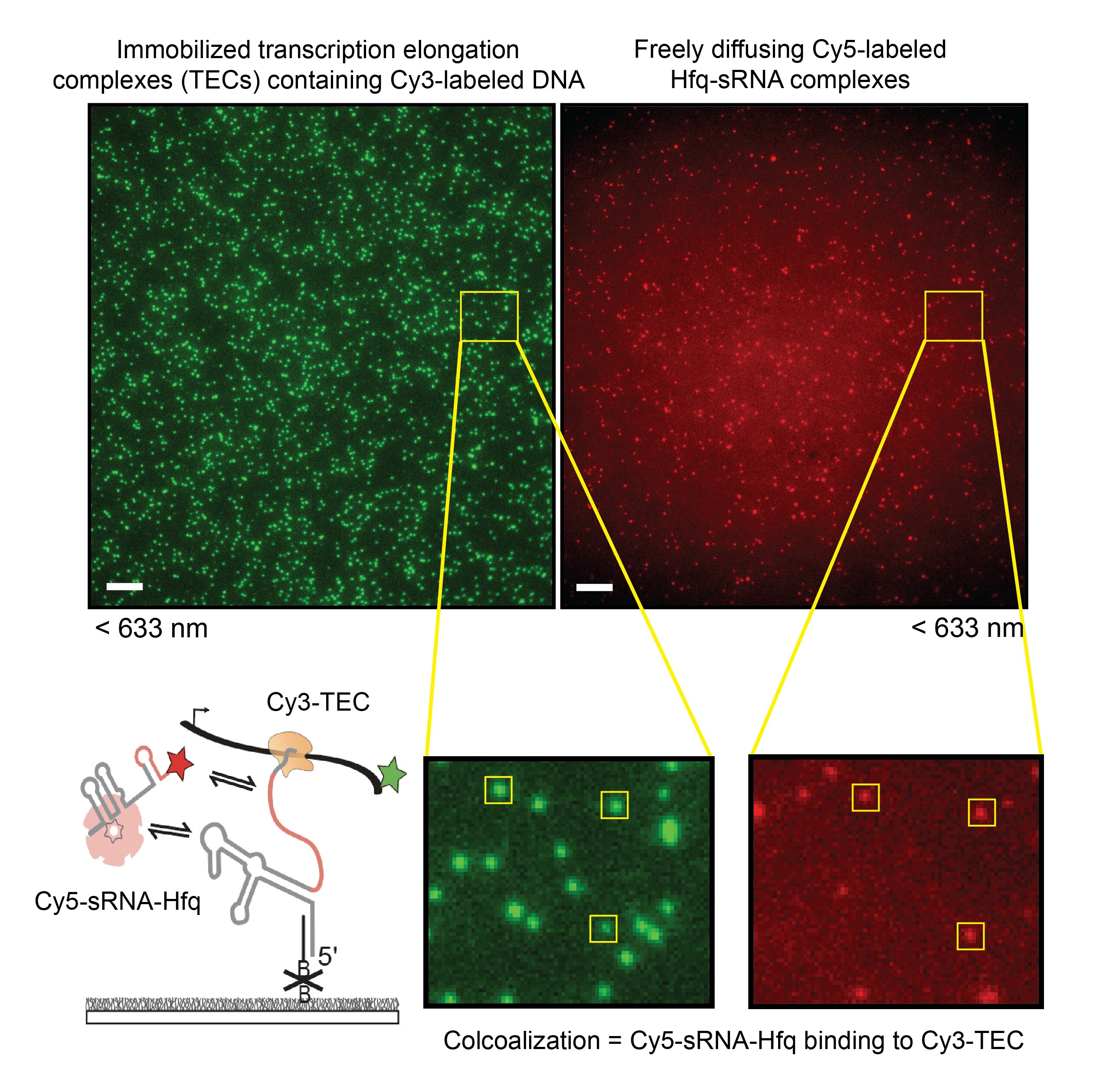
We aim to understand the molecular mechanisms that guide the assembly of RNPs in the cell. The lab focuses on studying co-transcriptional ribosome assembly as a lens to characterize the biochemical basis and biological consequences of coupling RNP assembly with transcription. Using biochemical and biophysical methods, like single-molecule fluorescence microscopy, we examine how RNAs fold during transcription and how protein binding influences transcription and RNA folding. We are also interested in studying many other RNPs including spliceosomes, microRNA/RNPs, messenger RNPs, and small nucleolar RNPs.
Research Images