About Our Research
The Section of Genetics and Physiology explores how cytokines control genetic programs, with an emphasis on the mammary gland, immune homeostasis and multiple autoimmune diseases (MAS). We use a range of genomic technologies to address two distinct, yet linked questions. First, we identify and investigate mutations that alter structure and function of proteins within the JAK-STAT signaling pathway, disrupt immune homeostasis and contribute to blood cancers and MAS. Secondly, we identify and investigate complex regulatory elements within the genome that control genetic programs executed by cytokines, such as interferons, interleukins and prolactin. We partner with community physicians and physicians Board Certified in Rheumatology, Laboratory Medicine and Infectious Diseases, with the goal to advance our knowledge of any role of JAK-STAT missense mutations in contributing to the female predominance of multiple autoimmune syndromes.
Current Research
Current research addresses critical barriers in understanding the impact of missense mutations in JAK-STAT components and related cytokine pathways in cellular homeostasis, blood cancers and autoimmune disease. Our research focuses on four topics:
- Identification and investigation of disease-associated missense mutation through partnerships with community-based research programs that include individuals diagnosed with autoimmune disease and their health providers.
- Utilization of community-derived databases optimized for capturing diversity, locate JAK-STAT mutations specifically linked to multiple immune syndrome (MAS), a relatively under-investigated topic, that would then represent the next targets of investigation in our laboratory.
- Impact of pregnancy hormones on kidney disease.
- Investigation of disease-associated SNPs causing missense mutations in the broader JAK-STAT pathway through experimental mouse genetics.
- Development of strategies and computational tools to investigate and predict the pathophysiological impact of SNPs in the human population (including ancient genomes).
References
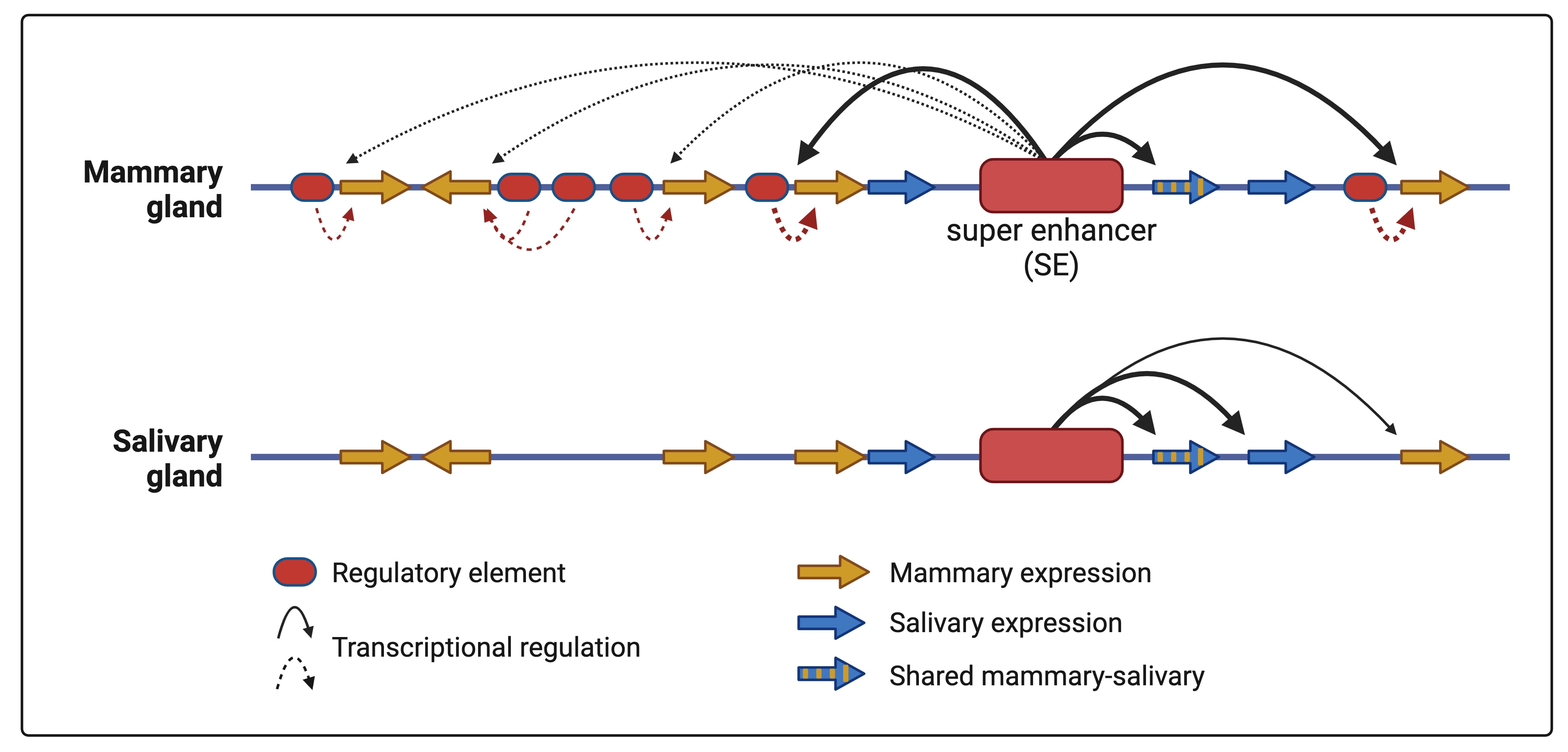
- Lee HK, Willi M, Liu C, Hennighausen L (2023) Cell-specific and shared enhancers control a multi-gene locus active in mammary and salivary glands. Nature Communications 14: 4992
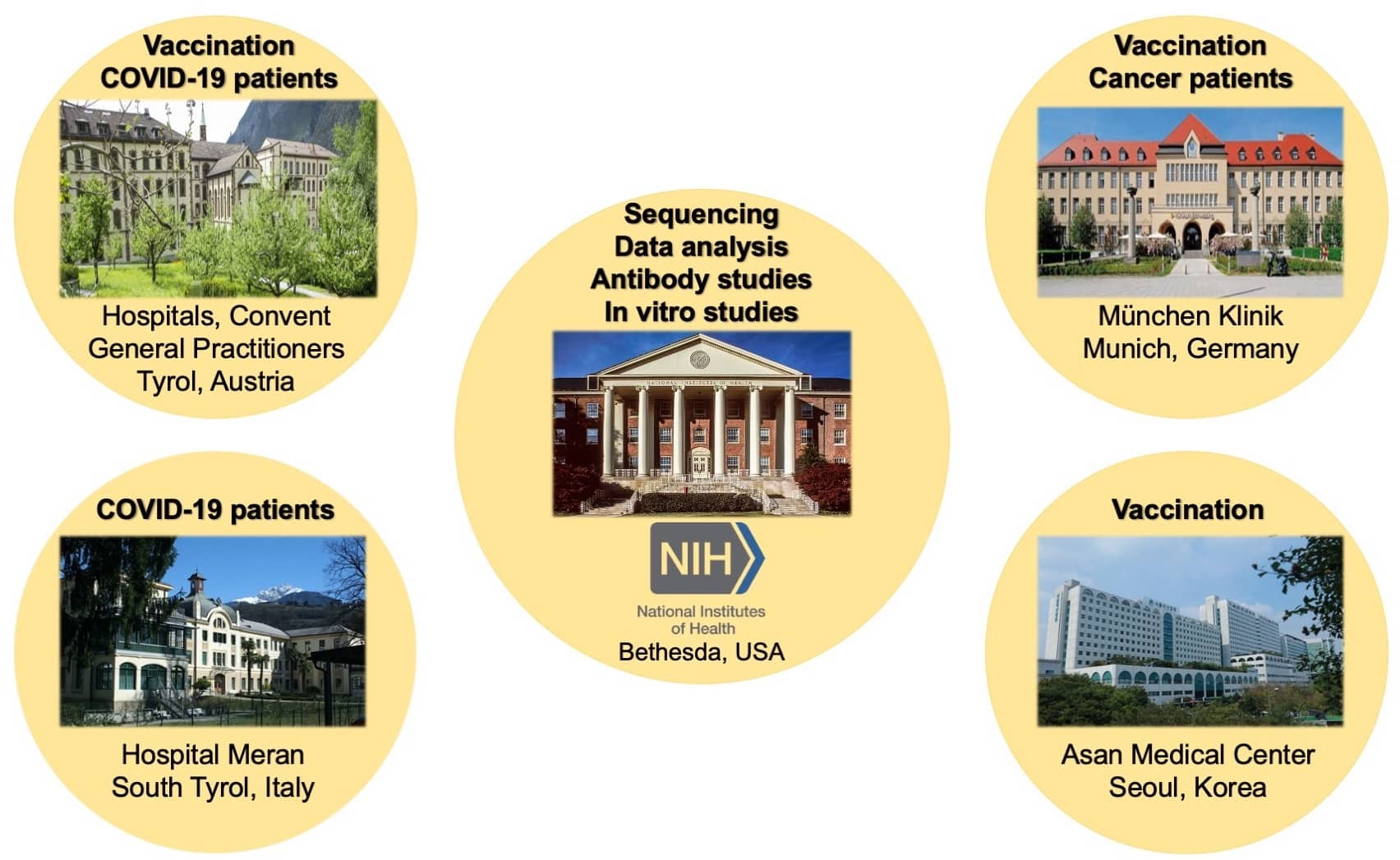
- Lee KH, Hoechstetter MA, Buchner M, Pham TT, Huh JW, Müller K, Zange S, von Buttlar H, Girl P, Wölfel R, Brandmeier L, Pfeuffer L, Furth PA, Wendtner CM, Hennighausen L (2023) Robust transcriptional response to COVID-19 immunization despite ineffective humoral immune responses in CLL patients. Blood Advances 7:2214-2227
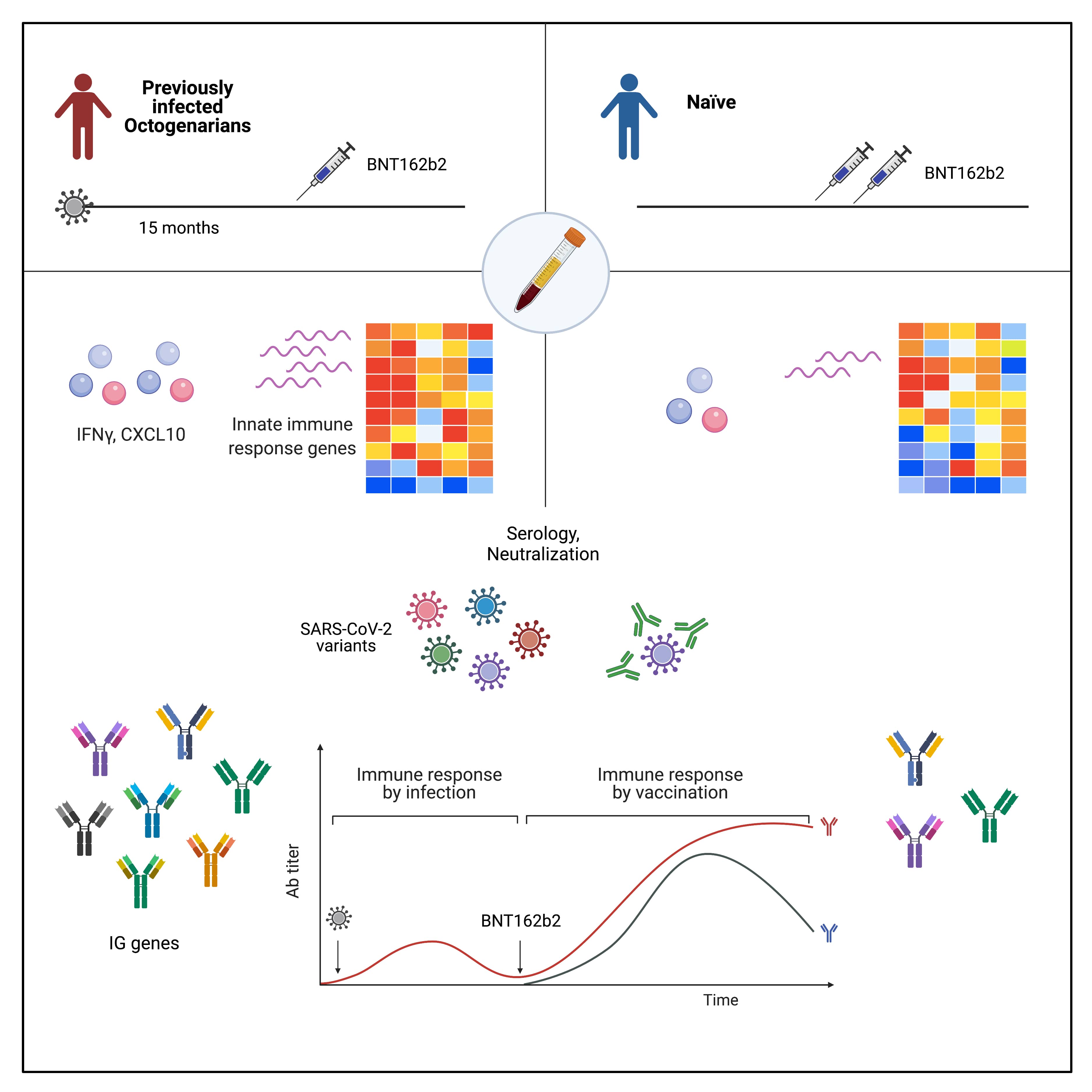
- Lee HK, Knabl L, Moliva JI, Knabl Sr. L, Werner AP, Boyoglu-Barnum S, Kapferer S, Pateter B, Walter M, Sullivan N, Furth PA, Hennighausen L (2022) mRNA vaccination in octogenarian nuns 15 and 20 months after recovery from COVID-19 elicits robust immune and antibody responses that include Omicron. Cell Reports 39:110680
- Lee HK, Knabl L, Pipperger L, Volland A, Furth PA, Smith HE, Knabl L, Bellmann R, Bernhard C, Kaiser N, Ganzer H, Strohle M, Walser A, von Laer D, Hennighausen L (2021) Immune transcriptomes of highly exposed SARS-CoV-2 asymptomatic seropositive versus seronegative individuals from the Ischgl community. Scientific Reports 11:4243
- Lee HK, Willi M, Kuhns T, Liu C, Hennighausen L (2021) Redundant and non-redundant cytokine-activated enhancers control Csn1s2b expression in the lactating mouse mammary gland. Nature Communications 12:2239
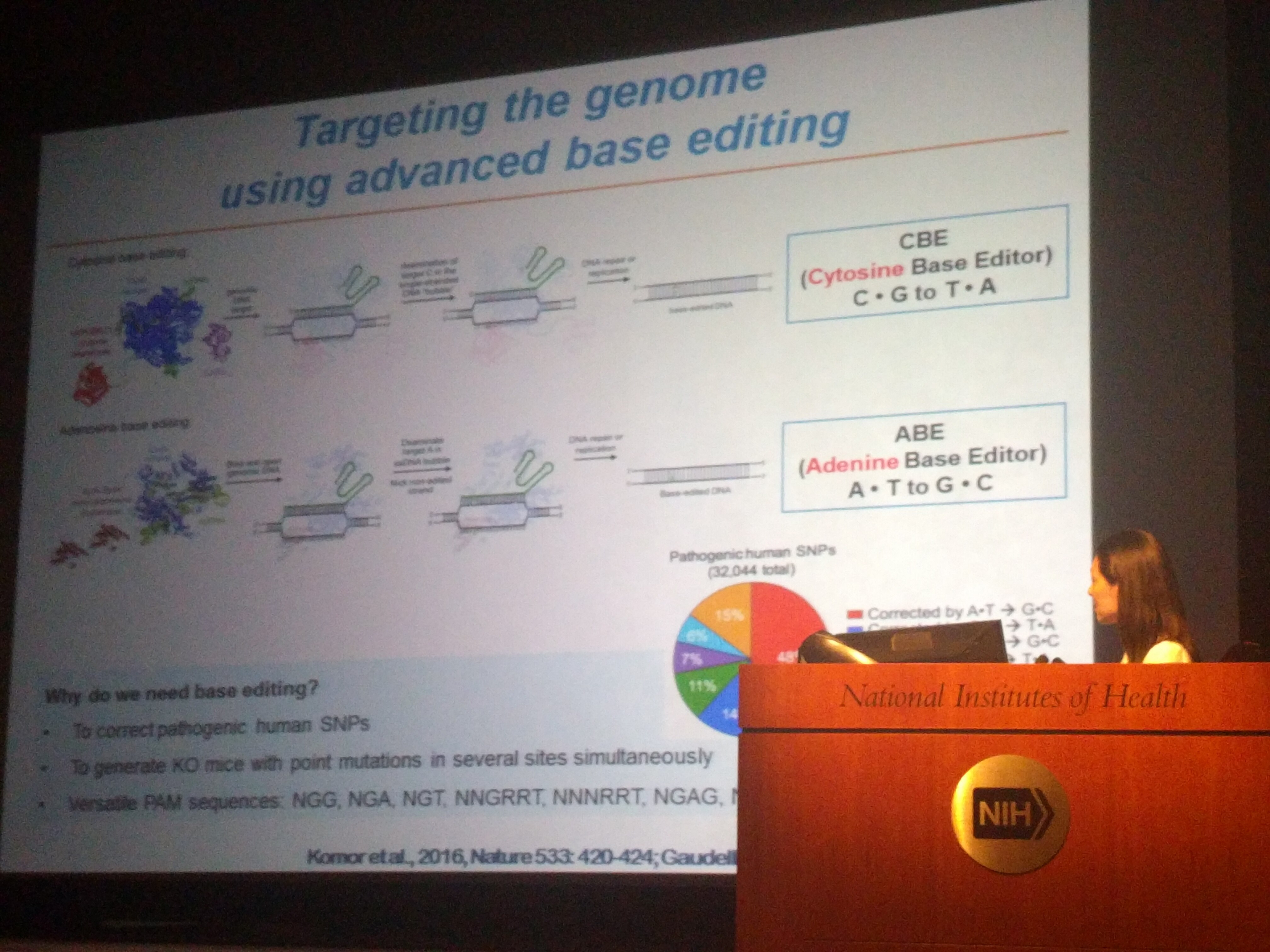
- Lee HK, Willi M, Miller SM, Kim S, Liu C, Liu DR, Hennighausen L (2018) Targeting fidelity of adenine and cytosine base editors in mice. Nature Communications 9:4804
- Lee HK, Willi M, Shin HY, Liu C, Hennighausen L (2018) Progressing super-enhancer landscape during mammary differentiation controls tissue-specific gene regulation. Nucleic Acids Res, 46: 10796-10809
- Shin HY, Wang C, Lee HK, Yoo KH, Zeng X, Kuhns T, Yang CM, Mohr T, Liu C, Hennighausen L (2017) CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nature Communications 8:15464
- Lee, H.K., Willi, M., Wang, C., Yang, C.M., Smith, H.E., Liu, C. and Hennighausen L. (2017) Functional assessment of CTCF sites at cytokine-sensing mammary enhancers using CRISPR/Cas9 gene editing in mice. Nucleic Acids Res, 45, 4606-4618
- Shin HY, Willi M, Yoo HK, Zeng X, Wang C, Metser G, Hennighausen L (2016) Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nature Genetics 48: 904-911
Current Collaborators
- Woo Kyun Bae (Hwasun Hospital, South Korea)
- So-young Bang (Hanyang University, Seoul, South Korea)
- Simon Choi (Chonnan National University, Gwangju, South Korea)
- Jin Won Huh (Asan Medical Center, Seoul, South Korea)
- Ludwig Knabl (Tyrol, Austria)
- Clemens Wendtner (Munich Germany)
- Markus List (Munich, Germany)
- Chengyu Liu (NHLBI, NIH)
- John O’Shea (NIAMS, NIH)
Our Section in the News
- NIH Catalyst: Creating a “Spellchecker” for Genes
- NIH I Am IRP Blog: Cutting-Edge Technique Simultaneously Edits Multiple Genetic Targets
- NIH Record: NIDDK Scientist Pedals His Way to 'Ancien' Status (PDF, 1.58 MB)
Research Images